Research Article - (2022) Volume 10, Issue 2
Performance evaluation and yield stability test of released food barley (Hordeum vulgare L.) varieties in highland areas of Siltie and Gurage Zones
Solomon Shibeshi* and Muluneh MekisoReceived: 05-May-2022, Manuscript No. AAFSF-22-59572; Editor assigned: 09-May-2022, Pre QC No. AAFSF-22-59572 (PQ); Reviewed: 22-May-2022, QC No. AAFSF-22-59572; Revised: 30-May-2022, Manuscript No. AAFSF-22-59572 (R); Published: 06-Jun-2022, DOI: 10.51268/2736-1799.22.10.075
Abstract
Improved barley varieties under barley production were limited in study areas. Therefore, testing of food barley varieties including check over locations for their performance is of paramount important in identification of superior varieties for production. The experiments were conducted to identify high yielding and to determine the stability of the varieties, and specifically and widely adapted food barley varieties for the target areas. Combined analysis of variance showed highly significant (p<0.05) differences among the varieties in plant height, spike length and grain yield, indicating the existence of a genetic difference of barley varieties. The highest mean grain yield (3506.80 kg ha-1) was obtained from variety Adosha and Robera (3279.20 kg ha-1) but HB-1307 which was check variety gave the relatively lowest mean grain yield (2465 ha-1). Regarding Wricke’s (Wi2), cultivar superiority measure (Pi) and CVi stability parameters, varieties Adosha and Tiret with lowest value were considered to be stable, showed wider adaptation and ranked best mean grain yield and were 1st and 4th, respectively; whereas HB-1307 and EH-1493 with higher value were unstable and shows specific adaptation. A large portion of the total sum of squares taken by genotype by environment interaction (GEI) (69.91%) shown there is vulnerability of grain yield to the influence of GEI. The highest yielding genotypes Robera and Adosha were winning over the environments; the existence of different winning genotypes in different environments confirmed the presence of crossover type of GEI. Therefore, the results suggested that the varieties Robera, Adosha and Cross-41/98 with other recommended management practices have to be promoted to the study areas and similar agro ecologies.
Keywords
Barley, Grain yield, Variety, Stability.
Introduction
Barley has been produced, since ancient times as staple food crops in the highlands of Ethiopia (MoA, 2019). It has great importance in social, food habit and malting purpose in the country. At the national level in 2015/16 cropping season, 944,401.34 ha of land is covered by food and malt barley and over 1856704.28 tons are produced. Major constraints affecting barely productivity are inadequacy of broadly adapted, high yielding, disease and insect resistant varieties; the weather conditions varying between seasons and locations; low level of crop management practices; the increasingly dwindling soil fertility situation, incidence of erratic diseases and insect pests, and escalation of climatic changes are growing concerns for barely production in Ethiopia. However, so far, studies on evaluation on food barley varieties in the study areas are limited (Hussein et al., 2000). Therefore, evaluating food barley varieties for their performance is a paramount important in identification of superior varieties for production. Hence the present study was initiated with the following objective: to identify high yielding and to determine the stability of the varieties, and identify specifically and widely adapted food barley varieties for the target areas (Eberhart and Russell, 1966).
Materials and Methods
Description of the study area
The experiment was conducted under rain-fed conditions during the 2019 to 2020 for two years (June to December). The experimental sites are representing the major agro-ecologies of two and six-row food barley growing areas. In these areas, ensete, barley, wheat, horse beans and field peas are the main food crops, in descending order of importance. Cabbage (kale) and carrot are the major cash crops in both zones (Mamo, 2017).
Experimental materials and design
In this experiment 11 nationally released food barley varieties were used. These were Robera, Cross-41/98, Adosha, EH-1493, Abdane, Gutaa, Tiret, Biftu, HB-1965, HB-1966 and HB-1307 were selected based on year of release, performance in previous trials and the agro- ecologies for which they were released.
The experimental layout used was randomized complete block design with three replications.
Seed was hand drilled on plot consisting of six 2.5 m long rows spaced at 0.20 m apart with a plot area of 3 m2. Spacing between blocks and plots was 1 m and 0.4 m, respectively. The middle four rows (2 m2) were used for data collection. Seed was sown at 125 kg ha-1, while fertilizers was applied uniformly at a rate of 38/19/7 kg ha-1 N/P2O5/S, respectively for all locations during sowing and urea was in split form half at sowing and stem elongation stages, respectively. Weed management and all other agronomic practices were carried out uniformly for all plots as required.
Data collection
Observations were made both on individual plants and plots in which the middle four rows were considered for plot and individual plant based data collection.
Tiller number per plant (TIPP): Was estimated from randomly taken area of 0.25 m2 (0.5 m × 0.5 m) by counting the number of plants after emergence and number of plants bearing fertile spike at maturity and considered their difference as tiller number.
Plant height (PH) (cm): Height of five randomly taken plants from each plot was measured from the ground surface to the base of the main stem spike and the average was recorded (Mekiso and Getahun, 2016).
Spike length (SL) (cm): Length of spikes from five randomly taken plants from each plot was measured from the base to tip of the spike and the average was recorded.
Number of kernels per spike (NKS): Were recorded from randomly selected five plants in the four middle rows from each plot after threshing counted by hand and averaged.
Biomass yield (BM) (kg ha-1): The total above ground biological yield (grain and straw), of the 4 middle rows was recorded for each plot after carefully sun drying the whole above ground harvested plants for three days (Francis and Kannenberg, 1978).
Thousand kernels weight (TKW) (gm): Was obtained by carefully counting 500 kernels samples from each plot and weighing on sophisticated sensitive balance after adjusted to the standard moisture content (12.50%) and then multiplying the result by two.
Grain yield (GY) (kg/ha): Was estimated by harvesting the middle four rows and adjusting the yield to standard moisture content 12.50% when the crop reached its physiological maturity and the average value was calculated and recorded.
Adjusted grain yield (kg/plot)= 100-GMC* (Yield per plot)
100-SMC (12.5%)
Where, GMC is moisture content of grain yield at harvest, SMC is standard moisture content.
Statistical data analysis
The data on 7 quantitative traits, including phenologic traits growth parameters plant height, spike length, tillers per plant, grain yield and yield components Number of kernels per spike, biomass yield, and thousand kernel weight were subjected to the analyses. Analysis of variance of data from combined over locations were conducted for all quantitative traits using SAS computer program using the models (Gomez and Gomez, 1984). The significance of differences among means was compared by DMRT (Duncan’s multiple range tests) at a probability level of 5%. The following RCBD ANOVA statistical model was used at each environment: Yij=μ+Gi+Bj+eij, Where, Yij=observed value of genotype i in block j, μ is the grand mean of the experiment, Gi=the effect of genotype i, Bj=the effect of block j, eij=error effect of genotype i in block j.
The model by Eberhart and Russell (1966), Yij=μi+βiIj+δij, define stability parameters that may be used to describe the performance of a variety over a series of environments. Yij is the varieties mean of the ith variety at the jth environment, μi is the ith variety means over all environments, βi is the regression coefficient that measures the response of the ith variety to varying environments, δij is the deviation from regression of the ith variety at the jth environment, Ij is the environmental index. Shukula stability variance (σi2) was computed, where varieties with minimum values are considered stable.
Stability was also measured by combining the mean yield and coefficient of variation (CVi) by Francis and Kannenberg‘s, 1978. Ecovalence (Wi2) suggested by Wricke (1962) and cultivar/environmental superiority measure (Pi) were computed to further describe stability. Statistical AMMI analysis of grain yield the AMMI model‘s IPCA1 and IPCA2 scores and GGE biplot for each variety were computed (Gauch and Zobel, 1997).
Results and Discussion
A significant variation was found among the food barley varieties in plant height, spike length, number of kernels, thousand kernel weights and grain yield at Alicho wuriro; plant height and grain yield at Endegagn and all parameters except number of tillers per plant at Geta district (Gauch and Zobel, 1988). The highest mean grain yield of the varieties Robera, Adosha and Abdane (3772.30, 3523.90 and 3160.70 kg ha-1) was obtained at Alicho wuriro district. The highest grain yield was obtained by variety Robera followed by Adosha have yield advantage of 83.17% and 71.10% over the check (HB-1307). The highest mean grain yield of the varieties HB-1966, EH-1493 and HB-1307 (3652.30, 3614.40 and 3592.00 kg ha-1) was obtained at Endegagn district. The highest grain yield was obtained by variety Robera followed by Adosha have yield advantage of 1.68% and 0.62% over the check (HB-1307). The highest mean grain yield of the varieties Cross -41/98, Adosha and Gutaa (4080.10, 3701.40 and 3632.40 kg ha-1) was obtained in Geta district. The highest grain yield was obtained by variety Cross-41/98 followed by Adosha have yield advantage of 134.06% and 112.30% over the check (HB-1307) (Table 1A).
| Code of variety | Alicho wuriro | Endegagn | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | SL | NTP | BM | KNS | TKW | GY | PH | SL | NTP | BM | KNS | TKW | GY | |
| G1 | 75.13abc | 7.25a-d | 3.33 | 9733.00 | 48cd | 43.03abc | 3772.3a | 91.7ab | 6.20 | 1.80 | 12133.0 | 37.44 | 28.58 | 2564.8d |
| G2 | 77.27ab | 6.57cd | 3.00 | 9733.00 | 51.5bcd | 49.67a | 2638.5abc | 89.3ab | 7.10 | 2.00 | 12133.0 | 42.44 | 43.70 | 2893.6cd |
| G3 | 75.63ab | 6.4d | 2.87 | 11067.00 | 50.5cd | 42.67a-d | 3523.9ab | 70.4c | 6.30 | 2.20 | 10533.0 | 49.89 | 26.52 | 3294.9abc |
| G4 | 69.8bcd | 7.58a-d | 2.53 | 11200.00 | 58.0abc | 39.3bcd | 2370.3bc | 94.6ab | 7.10 | 2.50 | 15733.0 | 49.22 | 44.08 | 3614.40a |
| G5 | 67.67cd | 7.9ab | 2.73 | 9067.00 | 46.17d | 40.42bcd | 3160.7abc | 97.33a | 6.40 | 2.20 | 10933.0 | 39.55 | 41.97 | 2963.8cd |
| G6 | 77.97a | 6.93bcd | 2.87 | 8533.00 | 52.67a-d | 43.12abc | 2779abc | 93.5ab | 6.20 | 1.90 | 10000.0 | 29.11 | 41.68 | 2977.3bcd |
| G7 | 76.63ab | 8.21a | 3.47 | 10267.00 | 48.83cd | 38.82bcd | 3091.9abc | 96.13a | 7.40 | 2.50 | 12000.0 | 42.11 | 47.42 | 3124.8a-d |
| G8 | 70.53a-d | 8.17a | 2.93 | 10667.00 | 5433a-d | 37.42cd | 2894.7abc | 95.6ab | 6.00 | 2.30 | 10133.0 | 38.55 | 28.77 | 3080.6a-d |
| G9 | 77.4ab | 7.23a-d | 4.27 | 11467.00 | 51.33a-d | 45.73ab | 2793.5abc | 86.4b | 6.70 | 2.00 | 11200.0 | 38.89 | 39.58 | 2838.0cd |
| G10 | 67.29d | 7.77abc | 3.10 | 9733.00 | 63.0a | 42.25bcd | 2557.3bc | 90.4ab | 6.30 | 2.60 | 15600.0 | 23.33 | 48.68 | 3652.3a |
| G11 | 53.57e | 7.4a-d | 2.60 | 6400.00 | 62.17ab | 35.45d | 2059.5c | 94.5ab | 6.40 | 3.00 | 15733.0 | 43.83 | 44.11 | 3592.0ab |
| Mean | 71.73 | 7.4 | 3.06 | 9806 | 53.36 | 41.62 | 2876.52 | 90.89 | 6.60 | 2.28 | 12376.0 | 39.49 | 39.55 | 3145.2 |
| CV% | 6.23 | 9.68 | 25.6 | 21.47 | 12.31 | 10.33 | 24.46 | 6.02 | 9.54 | 22.97 | 32.31 | 33.15 | 33.02 | 11.59 |
| LSD | 7.61** | 1.22* | 1.33ns | 3585.4ns | 11.19* | 7.32* | 1198.10* | 9.32** | 1.07ns | 0.89ns | 6809ns | 20.95ns | 22.25ns | 620.66** |
NOTE: Means with similar letters are not significantly (p<0.05) different, ** highly significant at 0.01 and * significant and ns=not significant at 0.05 probability levels; PH=Plant height (cm), SL=Spike length (cm), NTP=tillers per plant, NKS=Number of kernels per spike, BM=Biomass (kg/ha), TKW=Thousand kernel weight (g/plot), GY=Grain yield (kg/ha); G1(Robera), G2(Cross-41/98), G3(Adosha), G4(EH-1493), G5(Abdane), G6(Gutaa), G7(Tiret), G8(Biftu), G9(HB-1965), G10(HB-1966 and G11(HB-1307).
Plant height of the varieties was ranged from 53.57 (HB-1307) at Alicho wuriro to 97.33 cm (Abdane) at Alicho wuriro whereas thousand seed weight also ranged from 26.52 (Adosha) at Endegagn to 56.70 gm. Generally, Endegagn had better grand mean performance in grain yield for food barley production despite the need for testing across seasons (Table 1B).
| Code of variety | Geta | ||||||
|---|---|---|---|---|---|---|---|
| PH | SL | NTP | BM | KNS | TKW | GY | |
| G1 | 90.22a | 7.0a | 2.11 | 10133.0a | 50.17b | 54.3a | 3500.4abc |
| G2 | 88.44a | 7.0a | 2.11 | 11333.0a | 49.0bc | 54.63a | 4080.10a |
| G3 | 90.56a | 6.89a | 1.89 | 11067.0a | 55.67ab | 51.47a | 3701.4ab |
| G4 | 90.0a | 8.33a | 1.89 | 8533.0a | 61.17ab | 53.37a | 2610.8bcd |
| G5 | 86.33a | 7.67a | 2.22 | 8667.0a | 48.5bc | 47.73a | 2054.0d |
| G6 | 88.55a | 6.89a | 2.45 | 11467.0a | 67.67a | 56.3a | 3632.4abc |
| G7 | 90.33a | 8.0a | 2.45 | 11200.0a | 56.0ab | 53.17a | 3394.9abc |
| G8 | 90.89a | 8.0a | 2.22 | 10933.0a | 54.17ab | 55.27a | 2484.9cd |
| G9 | 84.11a | 6.78a | 2.33 | 10267.0a | 49.33bc | 56.7a | 3454.9abc |
| G10 | 92.78a | 8.22a | 2.33 | 10933.0a | 59.50ab | 53.77a | 2692.3bcd |
| G11 | 75.35b | 4.0b | 0.78 | 4133.0b | 35.5c | 33.10b | 1743.4d |
| Mean | 84.40 | 7.16 | 2.07 | 9878.80 | 53.33 | 51.80 | 3031.73 |
| CV% | 12.1 | 16.58 | 25.91 | 17.95 | 15.92 | 10.22 | 23.05 |
| LSD | 17.39* | 2.02** | 0.91ns | 3020.6** | 14.46* | 9.02** | 1190.3** |
NOTE: Means with similar letters are not significantly (p<0.05) different, ** highly significant at 0.01 and * significant and ns=not significant at 0.05 probability levels; PH=Plant height (cm), SL=Spike length (cm), NTP=tillers per plant, NKS=Number of kernels per spike, BM=Biomass (kg/ha), TKW=Thousand kernel weight (g/plot), GY=Grain yield (kg/ha); G1(Robera), G2(Cross-41/98), G3(Adosha), G4(EH-1493), G5(Abdane), G6(Gutaa), G7(Tiret), G8(Biftu), G9(HB-1965), G10(HB-1966 and G11(HB-1307).
In Table 2 indicated that combined analysis of variance showed highly significant (p<0.05) differences among the varieties in plant height, spike length and grain yield, indicating the existence of a genetic difference of barley varieties. The tallest mean plant height was recorded by Tiret (87.70 cm), while HB-1307 was the shortest (61.31 cm) which was below the grand mean among the tested varieties. Even though analysis of variance showed no significant difference among varieties, thousand kernel weights ranged from variety HB-1307 (37.54 gram) to Cross-41/98 (49.33 gram) was obtained, respectively. The highest mean grain yield (3506.80 kg ha-1) was obtained from variety Adosha followed by Robera (3279.20 kg ha-1) but HB-1307 which was check variety gave the relatively lowest mean grain yield (2465 ha-1) in the two cropping years. This result is supported by the finding of the same study which reported that varieties differ in grain yield (Teshome, 2017) (Tables 1-A and 1-B).
| Code of variety | PH (cm) | SL (cm) | NTP | BM (kg) | KNS | TKW (g) | GY (kg) |
|---|---|---|---|---|---|---|---|
| G1 | 85.70ab | 6.82bcd | 2.43 | 10667.00 | 45.20 | 41.97 | 3279.20ab |
| G2 | 84.99ab | 6.90bc | 2.37 | 11067.00 | 47.65 | 49.33 | 3204.10ab |
| G3 | 78.86b | 6.64cd | 2.32 | 10889.00 | 52.02 | 40.22 | 3506.80a |
| G4 | 84.80ab | 7.68ab | 2.32 | 11822.00 | 56.13 | 45.58 | 2865.20bc |
| G5 | 83.78ab | 7.31abc | 2.38 | 9556.00 | 44.74 | 43.37 | 2726.20bc |
| G6 | 86.66a | 6.69cd | 2.42 | 10000.00 | 49.81 | 47.03 | 3129.60ab |
| G7 | 87.70a | 7.88a | 2.81 | 11156.00 | 48.98 | 46.47 | 3203.90ab |
| G8 | 85.67ab | 7.39abc | 2.47 | 10578.00 | 49.02 | 40.48 | 2820.10bc |
| G9 | 82.64ab | 6.90bc | 2.87 | 10978.00 | 46.68 | 47.34 | 3028.60abc |
| G10 | 83.49ab | 7.44abc | 2.67 | 12089.00 | 48.61 | 48.23 | 2967.30abc |
| G11 | 61.31c | 5.93d | 2.13 | 8756.00 | 47.16 | 37.54 | 2465.00c |
| Mean | 82.33 | 7.05 | 2.47 | 10686.87 | 48.73 | 44.33 | 3017.80 |
| CV% | 8.71 | 12.47 | 25.34 | 26.22 | 19.35 | 19.19 | 20.21 |
| LSD | 6.76** | 0.83** | 0.59ns | 2642.60ns | 8.89ns | 8.02ns | 575.14** |
Stability analysis
After testing the significance of GEI mean square in combined ANOVA, stability analysis was conducted for grain yield using means of genotype in each location. SAS (Hussein et al., 2000) was used to conduct stability analysis (Table 2). Regarding Wricke’s (Wi2) stability parameter, varieties G7, G3 and G8 with lowest Wricke’s Eco valence were considered to be stable as they contribute 74826.92, 172839.70 and 186231.10% to the interaction sum of squares, showed wider adaptation and G3 and G7 were ranked best mean grain yield and were 1st and 4th, respectively; whereas G11, G1 and G2 with higher Wricke’s Eco valence value were unstable and made the highest contributions 1610214, 1153793 and 1114903% to GEI with higher average grain yield than grand mean except G11 and shows specific adaptation. However, cultivar superiority measure (Pi) depicted G3, G7 and G1 as stable, indicating wider adaptation across the environments; whereas G11, G5 and G4 were the most unstable varieties, respectively that showed limited adaptation. Parameter CVi, varieties G7, G3 and G8 were stable and had the lowest CVi, but higher grain yield than grand mean except G8, whereas G11, G2 and G4 with the highest CVi values had low yield performances, indicating unstable varieties. Accordingly varieties G3, G1, G2 and G7 the highest yielding varieties over all environments had linear regression coefficient of -0.73, -4.33, 1.45 and 0.23, respectively. Varieties with the lowest bi; G1, G5 and G3 were more adapted to marginal environments, whereas G11, G4 and G10 were input sensitive adapted to ideal environments for selecting varieties with specific adaptation (Table 3).
| Source | DF | SS | MS | Fpr | % SS (Var+Env+Var × ENV |
|---|---|---|---|---|---|
| Total | 98 | 66461452 | 678178 | ||
| Treatment | 32 | 39815037 | 1244220 | 3.34 | |
| Variety | 10 | 11830720 | 1183072 | 3.18 | 25.99 |
| Environment | 2 | 1298550 | 649275 | 0.90 | 4.10 |
| Block | 6 | 4325370 | 720895 | 1.94 | |
| Interactions | 20 | 26685768 | 1334288 | 3.59 | 69.91 |
| IPCA1 | 11 | 21163025 | 1923911 | 5.17 | 71.91 |
| IPCA2 | 9 | 5522743 | 613638 | 1.65 | 28.09 |
| Residuals | 0 | 0 | |||
| Error | 60 | 22321045 | 372017 |
AMMI analysis of GEI
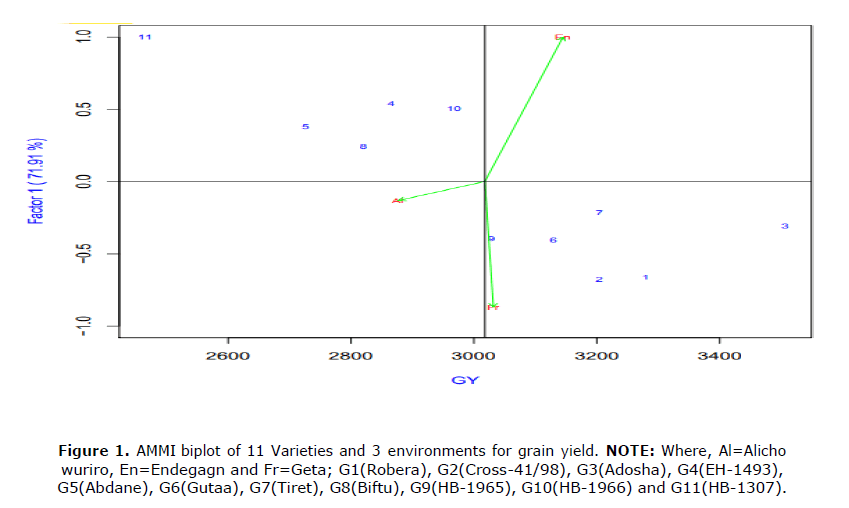
The proportion of sum of square of the treatment accounted for environment, genotype and GxE (Table 4). In the case of grain yield, the result showed that the variation explained by GEI was 69.91%, varieties took 25.99% and environment had shared 4.10% total sum of square. This result is in agreement with the findings of Gauch and Zobel (1997) who found significant differences among the genotypes, environment and GEI effects variation in barley grain yield. A large portion of the total sum of squares taken by GEI shown there is vulnerability of grain yield to the influence of GEI. The GEI was further partitioned by PCA. It is evident from Table 3 that using biplot in interactions was very limited where the first IPCA axes explain 71.91% of the total interaction. The partitioning of %SS indicated that the interaction effect was a predominant source of variation followed by the variety and environment effect. The variety effect was approximately six times higher than environment, which suggests the possible existence of different variety groups (Crossa et al., 1990) (Figure 1).
| Code of variety | Mean of yield kg/ha | Variation coefficient CVi (%) | Eberhart and Russell (bi) | Shuckla’s variance δ2 |
Wricke's Ecovalence (Wi) | Superiority measure (Pi) | Non parametric Nassar and Huehn | |
|---|---|---|---|---|---|---|---|---|
| Si(1) | Si(2) | |||||||
| G1 | 3279.15 | 19.32 | -4.33 | 667217 | 1153793 | 253104.3 | 5.67 | 26.5 |
| G2 | 3204.09 | 24.01 | 1.45 | 643450.8 | 1114903 | 310146 | 3 | 19 |
| G3 | 3506.77 | 5.81 | -0.73 | 67745.6 | 172839.7 | 55,460.95 | 1.33 | 1.5 |
| G4 | 2865.19 | 23.03 | 4.45 | 318823.3 | 583694 | 687599.7 | 4.67 | 17.5 |
| G5 | 2726.19 | 21.66 | -1.11 | 459864.0 | 814488 | 825503.2 | 2.33 | 13 |
| G6 | 3129.59 | 14.27 | 1.02 | 182750.3 | 361029.2 | 273753 | 1.33 | 5.5 |
| G7 | 3203.90 | 5.19 | 0.23 | 7848.9 | 74826.92 | 201,751.5 | 0.67 | 1 |
| G8 | 2820.08 | 10.81 | 0.49 | 75929.25 | 186231.1 | 606918.4 | 1.33 | 4.5 |
| G9 | 3028.60 | 12.20 | 0.41 | 132916.2 | 279482.5 | 335450.8 | 3 | 7 |
| G10 | 2967.28 | 20.12 | 3.89 | 247297.1 | 466651.2 | 567036.4 | 4.67 | 17.5 |
| G11 | 2464.95 | 40.11 | 5.25 | 946141.1 | 1610214 | 1399609 | 5.33 | 21.5 |
NOTE: Wi = Wricke’s ecovalence, (Pi) Lin and Binns’s cultivar performance measure, regression coefficient (bi), deviation from regression (S2d), CV = Coefficient Variability; G1(Robera), G2(Cross-41/98), G3(Adosha), G4(EH-1493), G5(Abdane), G6(Gutaa), G7(Tiret), G8(Biftu), G9(HB-1965), G10(HB-1966 and G11(HB-1307).
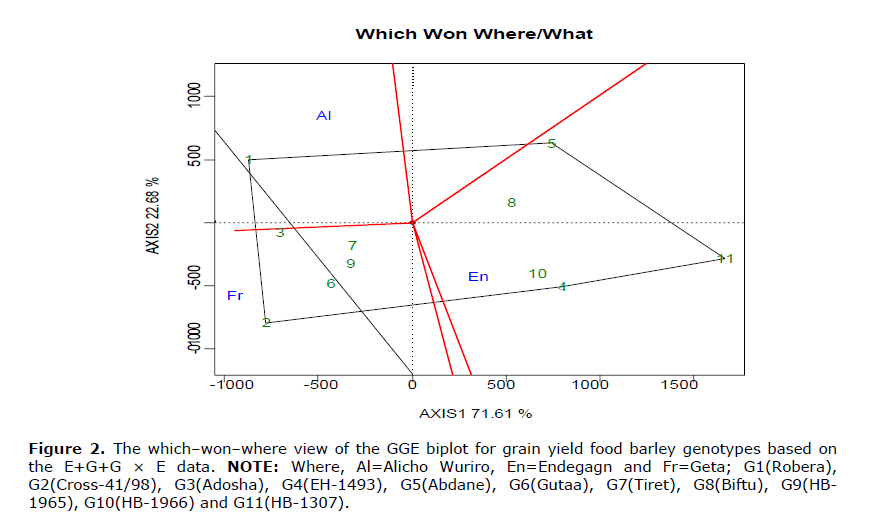
In the graph (Figure 2), a polygon is formed by connecting the genotypes that are on the vertex, while the other genotypes are contained in the polygon. A graph includes many rays divides the biplot into several sectors and the winning genotype for each sector is the one located on the respective vertex (Yan and Tinker, 2006). The highest yielding genotypes G2, G3, G1, G6, G9 and G7 were winning in the environment Geta, with in the vertex G2 and G1, while G10 and G4 with vertex G11 genotypes were the winning and high yielding once in environment Endegagn. The highest yielding genotypes G1, G3 and G5 were winning in the environment Alicho wuriro, with in the vertex G1. The existence of different winning genotypes in different environments confirmed the presence of crossover type of GEI.
Figure 2. The which–won–where view of the GGE biplot for grain yield food barley genotypes based on the E+G+G × E data. NOTE: Where, Al=Alicho Wuriro, En=Endegagn and Fr=Geta; G1(Robera), G2(Cross-41/98), G3(Adosha), G4(EH-1493), G5(Abdane), G6(Gutaa), G7(Tiret), G8(Biftu), G9(HB- 1965), G10(HB-1966) and G11(HB-1307).
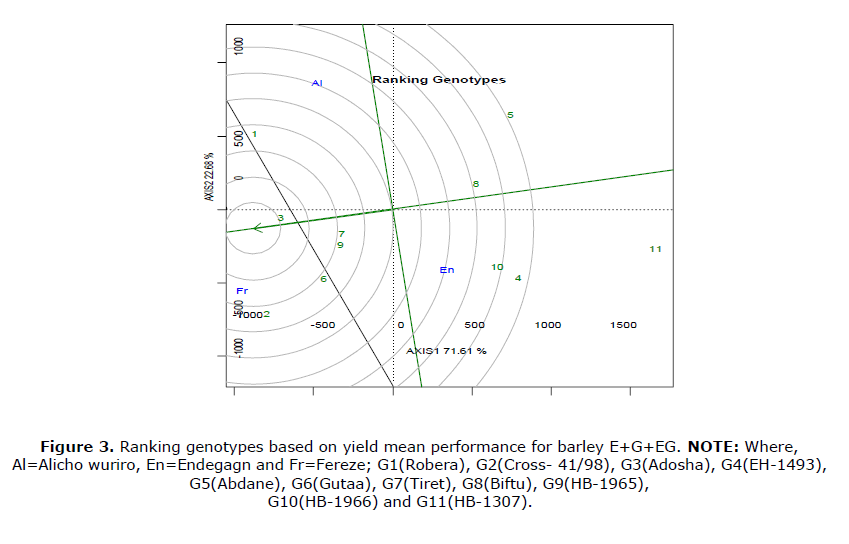
As shown in Figure 3, ranking genotypes relative to the ideal genotype is the use of GGE biplot. Genotypes found in the center of concentric circle on average environment coordinate x-axis designed to be equal to the longest vector of all genotypes and its projection on the average environment coordination y-axis was obviously zero that means it is absolutely stable (Wricke, 1962). Therefore, G3 is the ideal genotype (both stabile and high yielding) and G1, G6, G4, G7, G9 were the next ideal genotypes which were found near to Geta.
The GEI was further partitioned by PCA. It is evident from Table 3 that using biplot in interactions was very limited where the first IPCA axes explain 71.91 % of the total interaction. The partitioning of %SS indicated that the interaction effect was a predominant source of variation followed by the variety and environment effect.
The variety effect was approximately six times higher than environment, which suggests the possible existence of different variety groups (Crossa et al., 1990) (Figures 1-3).
In the graph Figure 2, a polygon is formed by connecting the genotypes that are on the vertex, while the other genotypes are contained in the polygon.
A graph includes many rays divides the biplot into several sectors and the winning genotype for each sector is the one located on the respective vertex (Yan W and Tinker B, 2006).
The highest yielding genotypes G2, G3, G1, G6, G9 and G7 were winning in the environment Geta, with in the vertex G2 and G1, while G10 and G4 with vertex G11 genotypes were the winning and high yielding once in environment Endegagn.
The highest yielding genotypes G1, G3 and G5 were winning in the environment Alicho wuriro, with in the vertex G1.
The existence of different winning genotypes in different environments confirmed the presence of crossover type of GEI. As shown in Figure 3, ranking genotypes relative to the ideal genotype is the use of GGE biplot.
Genotypes found in the center of concentric circle on average environment coordinate x-axis de- signed to be equal to the longest vector of all gen- otypes and its projection on the average environment coordination y-axis was obviously ze- ro that means it is absolutely stable (Wricke G, 1962).
Therefore, G3 is the ideal genotype (both stabile and high yielding) and G1, G6, G4, G7, G9 were the next ideal genotypes which were found near to Geta.
Conclusion
Adaptability of 11 improved food barley varieties (Robera, Cross-41/98, Adosha, EH-1493, Abdane, Gutaa, Tiret, Biftu, HB-1965, HB-1966 and HB- 1307as check) were evaluated at Alicho wuriro, Geta sub-testing site of Worabe Agricultural Re- search Centre and Endegagn district during 2019 and 2020 main cropping season.
Combined analysis of variance showed highly sig- nificant (p<0.05) differences among the varieties in plant height, spike length and grain yield, indi- cating the existence of a genetic difference of bar- ley varieties.
The highest mean grain yield (3506.80 kg ha-1) was obtained from variety Adosha and Robera (3279.20 kg ha-1) but HB-1307 which was check variety gave the relatively lowest mean grain yield (2465 ha-1).
A large portion of the total sum of squares taken by GEI shown there is vulnerability of grain yield to the influence of GEI. The highest yielding geno- types (G2, G3, G1, G6, G9 and G7) were winning in the environment Geta, with in the vertex G2 and G1, while G10 and G4 with vertex G11 geno- types were the winning and high yielding once in environment Endegagn. The highest yielding geno- types G1, G3 and G5 were winning in the envi- ronment Alicho wuriro, with in the vertex G1.
The existence of different winning genotypes in different environments confirmed the presence of crossover type of GEI.
Therefore, based on its agronomic performances observed from this experiment variety Robera, Adosha and Cross-41/98 could be popularized and scaled up to the farming communities of the test- ing area and to the farmers of similar agro ecolo- gy.
Future research actions should also focus on the performance of the selected three varieties with regard to the agronomic management and their traits.
Declaration
Author contributions: Conceptualization, SS; Methodology SS; Formal analysis, SS and MM; Investigation SS and MM, data curation, SS; Writing original draft preparation, SS and MM; Writing and editing SS and MM.
Funding
This research was funded by Southern Agricultural Research Institute.
Acknowledgment
The author acknowledges the Southern Agricul- tural Research Institute and Worabe Agricultural Research Centre for financing this study and providing the facilities required. The author is also indebted to the staff of Worabe Agricultural Re- search Centre in general, and to Mesfin Endrias, Daniel Kassa and Sadiya Abrar in particular, for their unreserved support in conducting the exper- iment.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the study design; in the col- lection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Author’s Statements
This research activity will help for food barely pro- ducer farmers and commercial producers to in- crease their wheat yield. The research result will also help for researcher who wants to do different research activities regarding food barely.
References
Eberhart SA, Russell WA (1966). Stability parameter for comparing varieties. CropScience. 6(1):36-40. [Crossref], [Google scholar]Francis TR, Kannenberg LW (1978). Yield stability studies in short-season maize. I. A descriptive method for grouping genotypes. Canadian Journal of Plant Sciences 58(4):1029-1034. [Crossref], [Google scholar]
Gauch HG, Zobel RW (1997). Identifying mega environments and targeting genotypes. Crop science. 37(2): 311-326. [Crossref], [Google scholar]
Gauch HG, Zobel RW (1988). Predictive and postdictive success of statistical analyses of yield trials. Theoretical and Applied genetics. 76(1):1-0. [Crossref], [Google scholar], [Indexed]
Gomez KA, Gomez AA (1984). Statistical Procedures for Agricultural Research, 2nd edition. The international rice research institute, Los Banos, Laguna, Philippinse. [Google scholar]
Hussein MA, Bjornstand A, Aastveit AH (2000). SAS program for Computing Genotype x Environment Stability Statistics. Agronomy Journal 92(3):454-459. [Crossref]
Mekiso M, Getahun M (2016). Area and Production of crops Private peasant holdings, Maher season. Agricultural Statistics. 1:14-28. [Google scholar]
MoA (Ministry of Agriculture) (2019). Status of Bovine Tuberculosis in Ethiopia: Challenges and Opportunities for Future Control and Prevention. National Livestock Development Project (NLDP) Working Paper 1-4, Addis Ababa, Ethiopia. [Google scholar]
Mamo T (2017) . Evaluation of improved food barley (Hordeum vulgare L.) varieties in the highland areas of Kaffa Zone, Southwestern Ethiopia. Agriculture, Forestry and Fisheries. 6(5):161-165. [Google scholar]
Wricke G (1962). Ubereine method zur Erfassung der Okologischen streuberite in Feldversuchen. Z. pflanzerzuentz. 47:92-96. [Google scholar]
Yan W, Tinker Bruce (2006). Application of GGE biplot analysis to evaluate genotype,environment and GxE interaction on P. radiata: a case study, University of New England, Armidale, NSW, 2351, Australia.